Contact information
Research groups
Colleges
Websites
-
NIHR
Funding body
-
Wellcome Trust
Funding body
-
MRC
Funding body
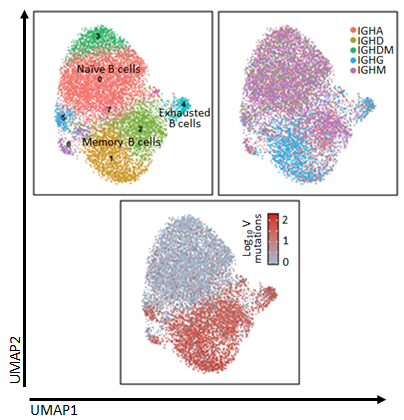
Figure 1 Single cell sequencing of B cells from NMDA receptor antibody encephalitis
Adam Handel
MRC New Investigator
- Associate Professor
- Consultant Neurologist
Current Research
I head the Oxford Autoimmune Neurology Group and lead a team of researchers investigating the role of different parts of the immune system in autoimmune conditions affecting the central nervous system (CNS).
My current research aims to understand clinical features and the molecular immunopathogenesis of neuroinflammatory conditions, with a particular focus on autoimmune encephalitis. I have identified clinical features characterizing several types of neurological conditions mediated by autoantibodies. I have generated high-throughput single-cell sequencing libraries from multiple tissues, including peripheral blood and cerebrospinal fluid, in patients with autoantibody-mediated CNS diseases (Fig. 1).
I have a particular interest in the role of T cells in autoimmune encephalitis. Although B cells have been extensively studied in autoimmune encephalitis, T cells have been relatively neglected. However, there are several strands of emerging evidence that T cells have a key role in this condition:
- One of the largest magnitude MHC associations in any autoimmune disease is found in LGI1 antibody encephalitis
- Patient derived autoantibodies are frequently immunoglobulin class switched and hypermutated, two characteristics which mandate T cell help and strongly implicate T cells as necessary for pathogenic autoantibody production
- There are marked T cell infiltrates within the brain parenchyma of patients with multiple types of autoimmune encephalitis
Therefore, T cells offer a novel immunotherapeutic target with translational potential that may address unmet clinical need in autoimmune encephalitis.
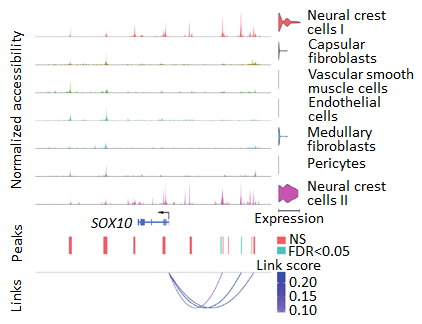
For autoreactive T cells to enter the peripheral immune repertoire, they must have escaped thymic negative selection, in which developing T cells are screened against multiple autoantigens by thymic epithelial cells and other stromal cell populations. To this end, my research also explores different aspects of thymic biology in both normal development and with genetic disruption of thymic epithelial cell function (Fig. 2). I used functional genomics methods to investigate the mechanisms underlying thymic function and other aspects of adaptive immunity.
Thus, through a combination of single cell sequencing and multiplexed spatial proteomics, I aim to understand how T cells with autoreactivity against CNS antigens can escape thymic negative selection.
I regularly teach undergraduate medical students at the University of Oxford and doctors on autoimmune encephalitis and functional neurological disorders.
Clinical role
I run the Oxford Autoimmune Neurology Clinic at the John Radcliffe Hospital with Oxford University Hospitals NHS Foundation Trust. In this capacity, I assess and treat patients with multiple different types of autoimmune encephalitis and other neuroinflammatory conditions. I am happy to be referred patients with autoimmune encephalitis from medical professionals throughout the UK.
Key publications
-
Immunotherapy‐Resistant Neuropathic Pain and Fatigue Predict Quality‐of‐Life in Contactin‐Associated Protein‐Like 2 Antibody Disease
Journal article
Ceronie B. et al, (2025), Annals of Neurology, 97, 521 - 528
-
The immunobiology of herpes simplex virus encephalitis and post-viral autoimmunity
Journal article
Cleaver J. et al, (2024), Brain, 147, 1130 - 1148
-
Cervical lymph nodes and ovarian teratomas as germinal centres in NMDA receptor-antibody encephalitis.
Journal article
Al-Diwani A. et al, (2022), Brain, 145, 2742 - 2754
-
Developmental dynamics of the neural crest-mesenchymal axis in creating the thymic microenvironment
Journal article
HANDEL A. et al, (2022), Science Advances
-
Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells
Journal article
Žuklys S. et al, (2016), Nature Immunology, 17, 1206 - 1215
Recent publications
-
Rethinking corticosteroid therapy in autoimmune neurology
Journal article
Chiek Teoh CS. et al, (2025), Brain
-
Clinical phenotype and outcomes in autoimmune encephalitis after herpes simplex virus encephalitis: a systematic review and meta-analysis.
Journal article
Cleaver J. et al, (2025), J Infect
-
Neurological examinations for psychiatric patients: A systematic review.
Journal article
Phillips J. et al, (2025), Schizophr Res, 284, 32 - 40
-
Thymic central tolerance takes centre stage in autoimmune disease
Journal article
Bunni SM-A. and Handel AE., (2025), The Lancet Regional Health - Western Pacific, 60, 101638 - 101638
-
Distinctive seizure signature in the first video case-control study of a naturally-occurring feline autoimmune encephalitis model.
Journal article
Binks SNM. et al, (2025), Brain Behav Immun, 126, 289 - 296
Figure 2 Single cell ATAC-RNA-seq multiomics in human thymus